New Year’s greetings and best wishes for 2020 to all!
We had ended last year, coincidentally, pondering why wide spread application of PCR-based tests in bacterial diagnostics has been elusive and identified a couple of key features that limit their use. It seems appropriate to start the new year with a discussion on one technology that can address those limitations – Infrared-based identification (IR).
What, you might be wondering, – that dilapidated instrument I vaguely recall using in undergraduate chemistry lab? Well, not quite that instrument but yes, one that utilizes the same fundamental principles.
The concept of using IR for identifying bacteria is not new. The earliest reference that we have been able to find dates back to 1950s! But, instrumental and computational limitations held back the potential for using this technology for identifying bacteria until the early 1980s. Naumann and colleagues took advantage of computational advances to demonstrate the ability of this technology to identify bacteria rapidly. Since then the use of IR for rapid and reliable identification of microbes to the strain level have been well documented. Reference databases to facilitate rapid identification of microorganisms are already available with some containing as many as 7000 strains.
How does the technology work?
IR, as you may recall, is a widely used vibrational spectroscopy technique that is used to identify compounds, even those present in mixtures. It is routinely used in raw material testing in the pharmaceutical and other industries as well as in testing of milk and milk products. All of the components that make up a bacterium (proteins, lipids, sugars, etc.) contribute to an IR “fingerprint” (see figure below). The nature and concentration of these components differ from one strain to another resulting in unique IR-fingerprints. This bacterial fingerprint has been used to differentiate between multiple bacterial species and strains including differentiating between antibiotic-resistant and -susceptible strains.
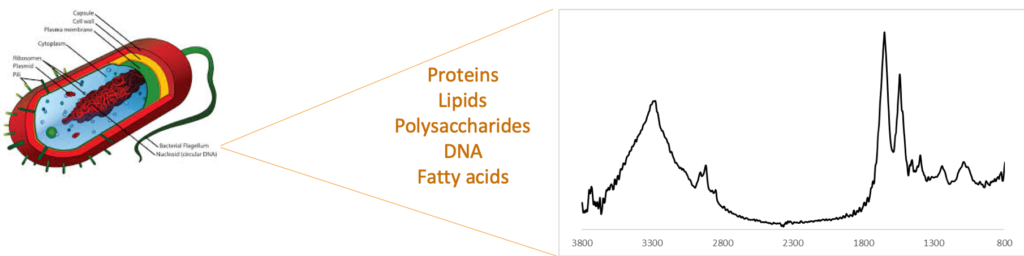
How does it compare to PCR-based identification?
Two key limitations of PCR-based tests that we had discussed previously were clinical sensitivity of the test and cost per test. If a PCR-test yielded a negative result, the clinician would not have any actionable information. On the other hand, with IR-based identification, the profile of a bacteria is very distinctive and its absence clearly indicates the absence of bacteria in the sample. In such a case, the clinician can actively consider whether administering antibiotics will be of any benefit to the patient. In the case of a positive result, the actions are similar to that with a PCR-based test with several important benefits. These are:
- the result is available in ≤ 10 minutes
- No custom labels or reagents are needed lowering the cost per test
- It does not require specialized laboratory for operation
- The bacteria is intact and viable after the analysis
- Antibiotic resistance can be determined
- The number of species and strains that are identified can be easily expanded without requiring new primers or antibiodies. This is done as a software update that consists of the fingerprint of the new bacteria and adjustments to the recognition algorithm, if needed.
You might be wondering at this stage why, if IR has all these benefits, there has not been any IR-based commercial instrument for use in bacterial ID? There are commercial instruments from Bruker, Thermo, and others that are used for bacterial ID in food safety testing. However the methodology used today requires bacteria to be cultured prior to identification (i.e. the same as most PCR-based tests). In addition, the bacteria has to be separated from the matrix components in order to make an accurate ID. Owing to these reasons, the technology has not yet seen widespread adoption for clinical use.
It is this aspect that we have impacted with the development of our separation cartridge. As mentioned previously, our separation cartridge can isolate and concentrate intact bacteria directly from blood with minimal manual intervention. The isolated bacteria can be identified using any technique. Using it upstream of IR however, permits rapid and sensitive identification of bacteria directly from patient sample without using any bacteria-specific labels or reagents. This combination of our separation cartridge and IR-based identification will, we believe, permit rapid, inexpensive, and hypothesis-free detection and identification. We aim to demonstrate this in 2020.
As we have said before, it’s not that IR (or any other ID technique) has to wholly replace PCR-based tests. The problem is large enough that multiple solutions will be needed. But, given the history of limited gains in developing effective PCR-based bacterial ID tests, the more diagnostic options we have, the better we can aid physicians in making the best decisions for treatment while slowing the spread of antimicrobial resistant bacteria. IR-based identification presents a compelling case to be a key diagnostic option.
